V.-Q. Vuong, C. Cevallos, B. Hourahine, B. Aradi, J. Jakowski, S. Irle, and C. Camacho
J. Chem. Phys. 158, 084802 (2023)
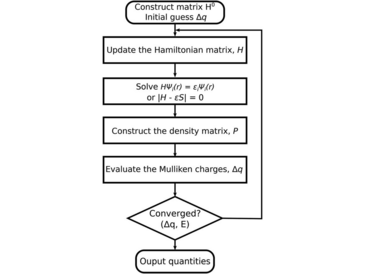
Acceleration of the density-functional tight-binding (DFTB) method on single and multiple graphical processing units (GPUs) was accomplished using the MAGMA linear algebra library. Two major computational bottlenecks of DFTB ground-state calculations were addressed in our implementation: the Hamiltonian matrix diagonalization and the density matrix construction. The code was implemented and benchmarked on two different computer systems: (1) the SUMMIT IBM Power9 supercomputer at the Oak Ridge National Laboratory Leadership Computing Facility with 1–6 NVIDIA Volta V100 GPUs per computer node and (2) an in-house Intel Xeon computer with 1–2 NVIDIA Tesla P100 GPUs. The performance and parallel scalability were measured for three molecular models of 1-, 2-, and 3-dimensional chemical systems, represented by carbon nanotubes, covalent organic frameworks, and water clusters.

![[Translate to English:]](/fileadmin/user_upload/fachbereiche/fb1/lmcqm/logos/lmcqm-logo-3-padded-up-20.png)