GFP-tagged bacteria endophytically in rice roots – Heatmap of differential gene expression – RNA-Seq
Research
Plants have evolved along with their associated microbes for millions of years, and they have high relevance for plant health, growth and productivity. Our research seeks to unravel these complex cooperation mechanisms, and also to bring them into use for a more sustainable agriculture.
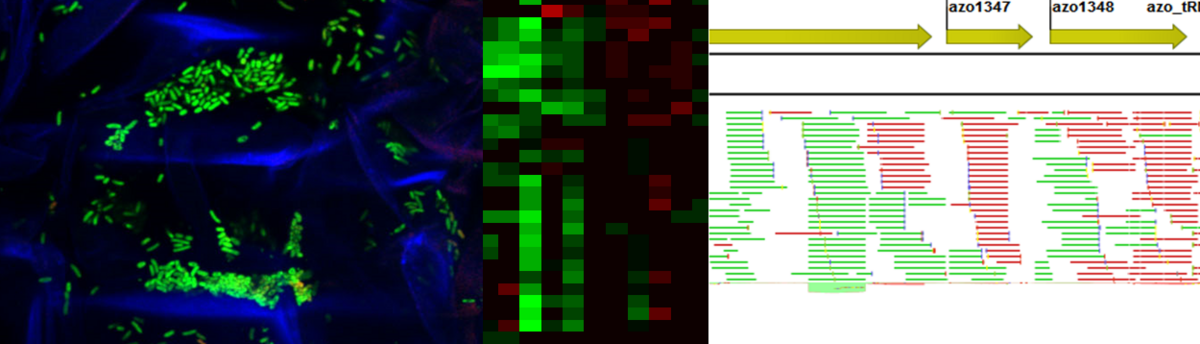
Molecular endophyte - plant interactions, Azoarcus olearius – rice model
Molecular endophyte - plant interactions: Azoarcus olearius – rice model
Endophytic bacteria are living inside plants, without causing symptoms of plant disease but rather being beneficial for the host. This life style in a rather specialized niche raises many questions about molecular mechanisms of cooperation, like: Which processes enable a successful establishment inside the plant? What are functions of bacteria inside the host?
As model system for root-bacteria-interactions, we study rice and the nitrogen-fixing endophyte Azoarcus olearius BH72, which expresses nitrogenase genes endophytically.
(Methods: Genomics, transcriptomics e.g. RNA-Seq, mutational analyses, synthetic biology, proteomics, qRT-PCR, light-, fluorescence-, electron and CLSM microscopy, rice transformation)

Molecular microbial ecology: Diversity and functions of root – associated bacteria
Rice in situ - Rice in phytotron and test tubes – Phylogenetic tree based on rRNA genes
Molecular microbial ecology: Diversity and functions of root – associated bacteria
Our knowledge of microbial diversity and function is still limited. Many microbes defy cultivation approaches in the laboratory. We seek to analyze their functional biodiversity in association with plants, by combining cultivation approaches and cultivation-independent state-of-the-art molecular tools. Polyphasic taxonomy allows us to characterize novel species. Synthetic communities consisting of of large collections of isolates from plants allow decomplexing the in situ system for better understanding the molecular basis of root microbiome functions. Microbial community analysis by sequencing or quantification of ribosomal marker genes, mRNAs, or metagenomics allows learning more about functions in situ, as in our CATCHY project.
(Methods: Classical and novel high-throughput isolation techniques, polyphasic taxonomy, molecular phylogeny, amplicon sequencing Illumina platform, q(RT)-PCR).

Rhizobial symbionts for sustainable agriculture in Africa
Cowpea field tests - Interactions with smallholders - Sampling - Active root nodules
Rhizobial symbionts for sustainable agriculture in Africa
Legume plants can gain their nitrogen demand by root nodule symbiosis with nitrogen-fixing rhizobia, and thus bring atmospheric nitrogen into crops and soil. An efficient symbiosis can benefit particularly smallholder farmers in Africa, enhancing productivity of pulses, improving the soil and providing protein-rich food. To contibute to sustainability of smallholder cropping systems in sub-Subsahara Africa, we identify and characterize well-adapted nodule symbionts for local pulses such as cowpea, Bambara groundnut, medicinal and undomesticated legumes (focus on Kavango region and Northern Namibia). Many of our isolates show a high temperature tolerance required for the harsh climate. Interaction with stakeholders and local partners will help to bring nitrogen-fixation into action. Previous projects The future Okavango (TFO) and SASSCAL, currently TOPSOIL and SUSTEC.
(Methods: Cultivation approaches, polyphasic taxonomy, genomics, small-scale inoculant production)
