Research
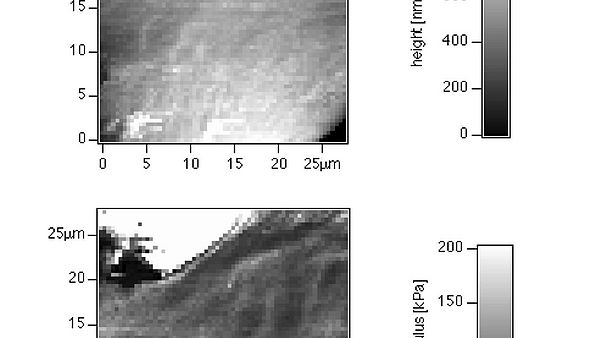
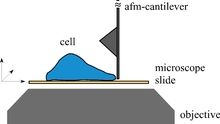
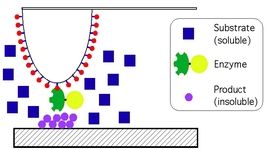
The research in the Radmacher lab focuses on the investigation of biological and biophysical processes. Examples are cell dynamics, cell mechanics, cell migration, cell division, activity of single enzyme molecules and enzymatic nanolithography. The most important tool for these investigations is the atomic force microscope (AFM). The AFM allows to investigate biological samples without any chemical fixation under physiological conditions combined with a high accuracy in positioning a very small tip and a high sensitivity in measuring and applying forces. This combination allows for instance to investigate biological processes from a cellular level down to single molecules. Although the AFM has been invented as an imaging device it has found recently several applications, where it is used to either to measure or to apply locally small forces on a sample. To learn more on AFM please click here.